Note
Go to the end to download the full example code
NWB File Basics¶
This example will focus on the basics of working with an NWBFile object,
including writing and reading of an NWB file, and giving you an introduction to the basic data types.
Before we dive into code showing how to use an NWBFile, we first provide
a brief overview of the basic concepts of NWB. If you are already familiar with the concepts of
TimeSeries and Processing Modules, then feel free to skip the Background: Basic concepts
part and go directly to The NWB file.
Background: Basic concepts¶
In the NWB Format, each experiment session is typically
represented by a separate NWB file. NWB files are represented in PyNWB by NWBFile
objects which provide functionality for creating and retrieving:
TimeSeries datasets, i.e., objects for storing time series data
Processing Modules, i.e., objects for storing and grouping analyses, and
experiment metadata and other metadata related to data provenance.
The following sections describe the TimeSeries and ProcessingModule
classes in further detail.
TimeSeries¶
TimeSeries objects store time series data and correspond to the TimeSeries specifications
provided by the NWB Format. Like the NWB specification, TimeSeries Python objects
follow an object-oriented inheritance pattern, i.e., the class TimeSeries
serves as the base class for all other TimeSeries types, such as,
ElectricalSeries, which itself may have further subtypes, e.g.,
SpikeEventSeries.
See also
For your reference, NWB defines the following main TimeSeries subtypes:
Extracellular electrophysiology:
ElectricalSeries,SpikeEventSeriesIntracellular electrophysiology:
PatchClampSeriesis the base type for all intracellular time series, which is further refined into subtypes depending on the type of recording:CurrentClampSeries,IZeroClampSeries,CurrentClampStimulusSeries,VoltageClampSeries,VoltageClampStimulusSeries.Optical physiology and imaging:
ImageSeriesis the base type for image recordings and is further refined by theImageMaskSeries,OpticalSeries, andTwoPhotonSeriestypes. Other related time series types are:IndexSeriesandRoiResponseSeries.Others
OptogeneticSeries,SpatialSeries,DecompositionSeries,AnnotationSeries,AbstractFeatureSeries, andIntervalSeries.
Processing Modules¶
Processing modules are objects that group together common analyses done during processing of data.
Processing module objects are unique collections of analysis results. To standardize the storage of
common analyses, NWB provides the concept of an NWBDataInterface, where the output of
common analyses are represented as objects that extend the NWBDataInterface class.
In most cases, you will not need to interact with the NWBDataInterface class directly.
More commonly, you will be creating instances of classes that extend this class.
See also
For your reference, NWB defines the following main analysis NWBDataInterface subtypes:
Behavior:
BehavioralEpochs,BehavioralEvents,BehavioralTimeSeries,CompassDirection,PupilTracking,Position,EyeTracking.Extracellular electrophysiology:
EventDetection,EventWaveform,FeatureExtraction,FilteredEphys,LFP.Optical physiology:
DfOverF,Fluorescence,ImageSegmentation,MotionCorrection.Others:
ImagingRetinotopy,Images.TimeSeries: Any TimeSeries is also a subclass of
NWBDataInterfaceand can be used anywhereNWBDataInterfaceis allowed.
Note
In addition to NWBContainer, which functions as a common base type for Group objects,
NWBData provides a common base for the specification of datasets in the NWB format.
NWB organizes data into different groups depending on the type of data. Groups can be thought of
as folders within the file. Here are some of the groups within an NWBFile and the types of
data they are intended to store:
acquisition: raw, acquired data that should never change
processing: processed data, typically the results of preprocessing algorithms and could change
analysis: results of data analysis
stimuli: stimuli used in the experiment (e.g., images, videos, light pulses)
The following examples will reference variables that may not be defined within the block they are used in. For clarity, we define them here:
from datetime import datetime
from uuid import uuid4
import numpy as np
from dateutil import tz
from pynwb import NWBHDF5IO, NWBFile, TimeSeries
from pynwb.behavior import Position, SpatialSeries
from pynwb.epoch import TimeIntervals
from pynwb.file import Subject
The NWB file¶
An NWBFile represents a single session of an experiment.
Each NWBFile must have a session description, identifier, and session start time.
Importantly, the session start time is the reference time for all timestamps in the file.
For instance, an event with a timestamp of 0 in the file means the event
occurred exactly at the session start time.
Create an NWBFile object with the required fields
(session_description, identifier,
session_start_time) and additional metadata.
Note
Use keyword arguments when constructing NWBFile objects.
session_start_time = datetime(2018, 4, 25, 2, 30, 3, tzinfo=tz.gettz("US/Pacific"))
nwbfile = NWBFile(
session_description="Mouse exploring an open field", # required
identifier=str(uuid4()), # required
session_start_time=session_start_time, # required
session_id="session_1234", # optional
experimenter=[
"Baggins, Bilbo",
], # optional
lab="Bag End Laboratory", # optional
institution="University of My Institution", # optional
experiment_description="I went on an adventure to reclaim vast treasures.", # optional
related_publications="DOI:10.1016/j.neuron.2016.12.011", # optional
)
nwbfile
Note
See the NWBFile Best Practices
for detailed information about the arguments to
NWBFile
Subject Information¶
In the Subject object we can store information about the experiment subject,
such as age, species, genotype, sex, and a description.
The fields in the Subject object are all free-form text (any format will be valid),
however it is recommended to follow particular conventions to help software tools interpret the data:
age: ISO 8601 Duration format, e.g.,
"P90D"for 90 days oldspecies: The formal Latin binomial nomenclature, e.g.,
"Mus musculus","Homo sapiens"sex: Single letter abbreviation, e.g.,
"F"(female),"M"(male),"U"(unknown), and"O"(other)
Add the subject information to the NWBFile
by setting the subject field to the new Subject object.
subject = Subject(
subject_id="001",
age="P90D",
description="mouse 5",
species="Mus musculus",
sex="M",
)
nwbfile.subject = subject
subject
Time Series Data¶
TimeSeries is a common base class for measurements sampled over time,
and provides fields for data and timestamps (regularly or irregularly sampled).
You will also need to supply the name and unit of measurement
(SI unit).

For instance, we can store a TimeSeries data where recording started
0.0 seconds after start_time and sampled every second:
data = list(range(100, 200, 10))
time_series_with_rate = TimeSeries(
name="test_timeseries",
data=data,
unit="m",
starting_time=0.0,
rate=1.0,
)
time_series_with_rate
For irregularly sampled recordings, we need to provide the timestamps for the data:
timestamps = list(range(10))
time_series_with_timestamps = TimeSeries(
name="test_timeseries",
data=data,
unit="m",
timestamps=timestamps,
)
time_series_with_timestamps
TimeSeries objects can be added directly to NWBFile using:
NWBFile.add_acquisitionto add acquisition data (raw, acquired data that should never change),NWBFile.add_stimulusto add stimulus data, orNWBFile.add_stimulus_templateto store stimulus templates.
nwbfile.add_acquisition(time_series_with_timestamps)
We can access the TimeSeries object 'test_timeseries'
in NWBFile from acquisition:
nwbfile.acquisition["test_timeseries"]
or using the method NWBFile.get_acquisition:
nwbfile.get_acquisition("test_timeseries")
Spatial Series and Position¶
SpatialSeries is a subclass of TimeSeries
that represents the spatial position of an animal over time.
Create a SpatialSeries object named "SpatialSeries" with some fake data.
# create fake data with shape (50, 2)
# the first dimension should always represent time
position_data = np.array([np.linspace(0, 10, 50), np.linspace(0, 8, 50)]).T
position_timestamps = np.linspace(0, 50).astype(float) / 200
spatial_series_obj = SpatialSeries(
name="SpatialSeries",
description="(x,y) position in open field",
data=position_data,
timestamps=position_timestamps,
reference_frame="(0,0) is bottom left corner",
)
spatial_series_obj
To help data analysis and visualization tools know that this SpatialSeries object
represents the position of the subject, store the SpatialSeries object inside
of a Position object, which can hold one or more SpatialSeries
objects.
Create a Position object named "Position" [1].
# name is set to "Position" by default
position_obj = Position(spatial_series=spatial_series_obj)
position_obj
Behavior Processing Module¶
ProcessingModule is a container for data interfaces that are related to a particular
processing workflow. NWB differentiates between raw, acquired data (acquisition), which should never change,
and processed data (processing), which are the results of preprocessing algorithms and could change.
Processing modules can be thought of as folders within the file for storing the related processed data.
Tip
Use the NWB schema module names as processing module names where appropriate.
These are: "behavior", "ecephys", "icephys", "ophys", "ogen", "retinotopy", and "misc".
Let’s assume that the subject’s position was computed from a video tracking algorithm, so it would be classified as processed data.
Create a processing module called "behavior" for storing behavioral data in the NWBFile
and add the Position object to the processing module using the method
NWBFile.create_processing_module:
behavior_module = nwbfile.create_processing_module(
name="behavior", description="processed behavioral data"
)
behavior_module.add(position_obj)
behavior_module
Once the behavior processing module is added to the NWBFile,
you can access it with:
nwbfile.processing["behavior"]
Writing an NWB file¶
NWB I/O is carried out using the NWBHDF5IO class [2]. This class is responsible
for mapping an NWBFile object into HDF5 according to the NWB schema.
To write an NWBFile, use the write method.
/home/docs/checkouts/readthedocs.org/user_builds/pynwb/envs/stable/lib/python3.11/site-packages/hdmf/build/objectmapper.py:260: DtypeConversionWarning: Spec 'TimeSeries/timestamps': Value with data type int64 is being converted to data type float64 as specified.
warnings.warn(full_warning_msg, DtypeConversionWarning)
You can also use NWBHDF5IO as a context manager:
Reading an NWB file¶
As with writing, reading is also carried out using the NWBHDF5IO class.
To read the NWB file we just wrote, use another NWBHDF5IO object,
and use the read method to retrieve an
NWBFile object.
Data arrays are read passively from the file.
Accessing the data attribute of the TimeSeries object
does not read the data values, but presents an HDF5 object that can be indexed to read data.
You can use the [:] operator to read the entire data array into memory.
test_timeseries pynwb.base.TimeSeries at 0x140204131361104
Fields:
comments: no comments
conversion: 1.0
data: <HDF5 dataset "data": shape (10,), type "<i8">
description: no description
interval: 1
offset: 0.0
resolution: -1.0
timestamps: <HDF5 dataset "timestamps": shape (10,), type "<f8">
timestamps_unit: seconds
unit: m
[100 110 120 130 140 150 160 170 180 190]
It is often preferable to read only a portion of the data.
To do this, index or slice into the data attribute just like you
index or slice a numpy array.
[100 110]
Note
If you use NWBHDF5IO as a context manager during read,
be aware that the NWBHDF5IO gets closed and when the
context completes and the data will not be available outside of the
context manager [3].
Accessing data¶
We can also access the SpatialSeries data by referencing the names
of the objects in the hierarchy that contain it. We can access a processing module by indexing
nwbfile.processing with the name of the processing module, "behavior".
Then, we can access the Position object inside of the "behavior"
processing module by indexing it with the name of the Position object,
"Position".
Finally, we can access the SpatialSeries object inside of the
Position object by indexing it with the name of the
SpatialSeries object, "SpatialSeries".
behavior pynwb.base.ProcessingModule at 0x140204131356880
Fields:
data_interfaces: {
Position <class 'pynwb.behavior.Position'>
}
description: processed behavioral data
Position pynwb.behavior.Position at 0x140204131359696
Fields:
spatial_series: {
SpatialSeries <class 'pynwb.behavior.SpatialSeries'>
}
SpatialSeries pynwb.behavior.SpatialSeries at 0x140204135431056
Fields:
comments: no comments
conversion: 1.0
data: <HDF5 dataset "data": shape (50, 2), type "<f8">
description: (x,y) position in open field
interval: 1
offset: 0.0
reference_frame: (0,0) is bottom left corner
resolution: -1.0
timestamps: <HDF5 dataset "timestamps": shape (50,), type "<f8">
timestamps_unit: seconds
unit: meters
Reusing timestamps¶
When working with multi-modal data, it can be convenient and efficient to store timestamps once and associate multiple
data with the single timestamps instance. PyNWB enables this by letting you reuse timestamps across
TimeSeries objects. To reuse a TimeSeries timestamps in a new
TimeSeries, pass the existing TimeSeries as the new
TimeSeries, pass the existing TimeSeries as the new
TimeSeries timestamps:
data = list(range(101, 201, 10))
reuse_ts = TimeSeries(
name="reusing_timeseries",
data=data,
unit="SIunit",
timestamps=time_series_with_timestamps,
)
Time Intervals¶
The following provides a brief introduction to managing annotations in time via
TimeIntervals. See the Annotating Time Intervals tutorial
for a more detailed introduction to TimeIntervals.
Trials¶
Trials are stored in TimeIntervals, which is
a subclass of DynamicTable.
DynamicTable is used to store
tabular metadata throughout NWB, including trials, electrodes and sorted units. This
class offers flexibility for tabular data by allowing required columns, optional
columns, and custom columns which are not defined in the standard.
The trials TimeIntervals class can be thought of
as a table with this structure:
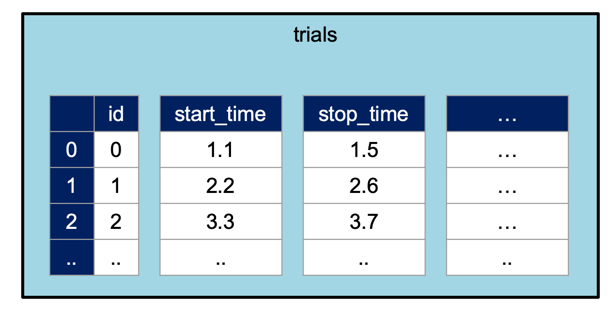
By default, TimeIntervals objects only require start_time
and stop_time of each trial. Additional columns can be added using
the method NWBFile.add_trial_column. When all the desired custom columns
have been defined, use the NWBFile.add_trial method to add each row.
In this case, we will add one custom column to the trials table named “correct”
which will take a boolean array, then add two trials as rows of the table.
nwbfile.add_trial_column(
name="correct",
description="whether the trial was correct",
)
nwbfile.add_trial(start_time=1.0, stop_time=5.0, correct=True)
nwbfile.add_trial(start_time=6.0, stop_time=10.0, correct=False)
DynamicTable and its subclasses can be converted to a pandas
DataFrame for convenient analysis using to_dataframe.
nwbfile.trials.to_dataframe()
Epochs¶
Like trials, epochs can be added to an NWB file using the methods
NWBFile.add_epoch_column and NWBFile.add_epoch.
The third argument is one or more tags for labeling the epoch, and the fourth argument is a
list of all the TimeSeries that the epoch applies
to.
nwbfile.add_epoch(
start_time=2.0,
stop_time=4.0,
tags=["first", "example"],
timeseries=[time_series_with_timestamps],
)
nwbfile.add_epoch(
start_time=6.0,
stop_time=8.0,
tags=["second", "example"],
timeseries=[time_series_with_timestamps],
)
nwbfile.epochs.to_dataframe()
Other time intervals¶
These TimeIntervals objects are stored in NWBFile.intervals. In addition to the default
epochs and trials, you can also add your own with custom names.
sleep_stages = TimeIntervals(
name="sleep_stages",
description="intervals for each sleep stage as determined by EEG",
)
sleep_stages.add_column(name="stage", description="stage of sleep")
sleep_stages.add_column(name="confidence", description="confidence in stage (0-1)")
sleep_stages.add_row(start_time=0.3, stop_time=0.5, stage=1, confidence=0.5)
sleep_stages.add_row(start_time=0.7, stop_time=0.9, stage=2, confidence=0.99)
sleep_stages.add_row(start_time=1.3, stop_time=3.0, stage=3, confidence=0.7)
nwbfile.add_time_intervals(sleep_stages)
sleep_stages.to_dataframe()
Now we overwrite the file with all of the data
/home/docs/checkouts/readthedocs.org/user_builds/pynwb/envs/stable/lib/python3.11/site-packages/hdmf/build/objectmapper.py:260: DtypeConversionWarning: Spec 'TimeSeries/timestamps': Value with data type int64 is being converted to data type float64 as specified.
warnings.warn(full_warning_msg, DtypeConversionWarning)
Appending to an NWB file¶
To append to a file, read it with NWBHDF5IO and set the mode argument to 'a'.
After you have read the file, you can add [4] new data to it using the standard write/add functionality demonstrated
above. Let’s see how this works by adding another TimeSeries to acquisition.
io = NWBHDF5IO("basics_tutorial.nwb", mode="a")
nwbfile = io.read()
new_time_series = TimeSeries(
name="new_time_series",
data=[1, 2, 3, 4, 5, 6, 7, 8, 9, 10],
timestamps=[1, 2, 3, 4, 5, 6, 7, 8, 9, 10],
unit="n.a.",
)
nwbfile.add_acquisition(new_time_series)
Finally, write the changes back to the file and close it.